3D Visualization and Gestural Interaction with Multimodal Neurological Data
Project team: J. Cooperstock (McGill ECE), T. Arbel (McGill ECE),
L. Collins (McGill MNI), R. Balakrishnan (Toronto CS),
R. Del Maestro (McGill Neurology and Neurosurgery), B. Goulet (McGill MUHC).
Funded as an NSERC Strategic Project.
Overview
This project deals with the challenges of medical image visualization, in particular within the domain of neurosurgery. We aim to provide effective ways of visualizing and interacting with brain data for training, planning, and surgical tasks.
Objectives include:
- Advanced scientific visualization
- Recognition of a usable set of input gestures for navigation/control
- Real-time communication of the data between multiple participants
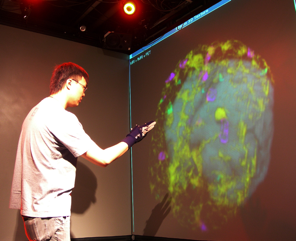
Efficient, contact-free manipulation coupled with multimodal data visualization would significantly enhance surgical training and procedures, and enable remote education and coaching.
Existing tools typically provide 2D slices and limited 3D surface renderings. True real-time manipulation of lighting, opacity, and viewpoint greatly improves comprehension. Current systems remain limited in computational intelligence and collaboration support.
Project Plan
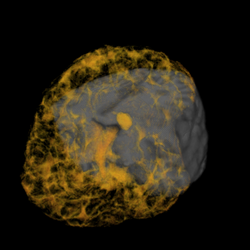
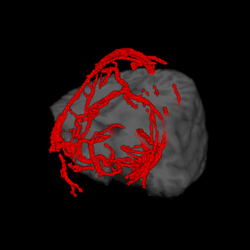

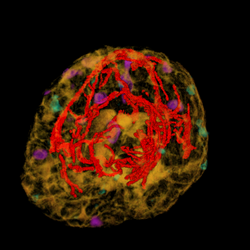
We are developing techniques for visualization and navigation of neurological data, with emphasis on distributed collaboration.
A major task is implementing an abstraction layer for rendering synthetic data, independent of display technology. This may leverage the IBIS system or custom rendering. The aim is to integrate MRI, fMRI, PET, DTI, and ultrasound with tool tracking and interactive manipulation.
Studies will evaluate prototype display/interaction paradigms with medical students, analyzing accuracy and performance in surgical tasks:
- Selective amygdala-hippocampectomy
- Depth electrode implantation (path planning)
- Tumor resections
- Spinal instrumentation (screw placement accuracy)
Future directions include gesture-based freehand tracking, augmented reality overlays, and peel-away visualization of tissue layers.
Virtual Brain Biopsy
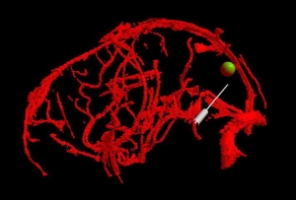




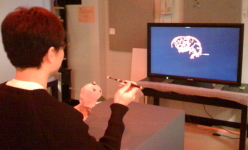
Task
Participants defined a straight vessel-free path from cortical
surface to tumor using a probe. The vasculature and tumor were
visualized with tangible props (plastic skull, biopsy probe).
Foot pedal input prevented probe jitter.
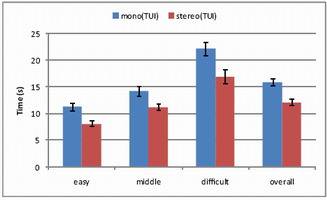
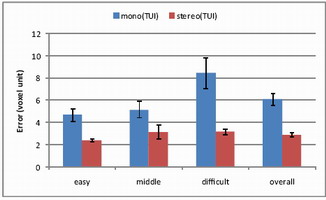
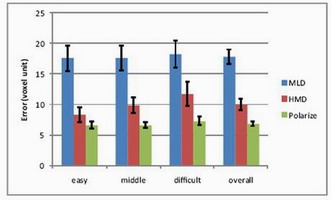
Results
Completion time and error (distance from probe tip to tumor center)
were measured across difficulty levels.
Mono vs. Stereo
With 12 participants (ages 22–37), stereo significantly reduced
completion time (p<0.0001) and error (p<0.0001).
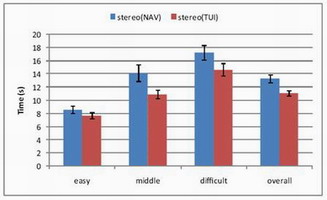
TUI vs. 3D Mouse
Compared tangible user interface vs. SpaceNavigator with 12
participants (ages 22–57).
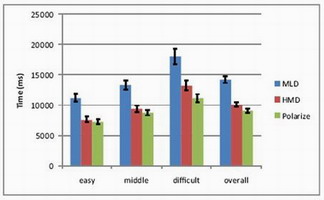
HMD vs. Polarized vs. MLD
Head-mounted display, polarized projection, and multiview lenticular
display were compared with 12 participants (ages 23–36). Polarized
projection achieved fastest completion and smallest error.
Medical Volume Carving
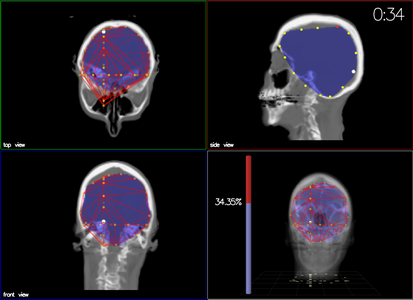
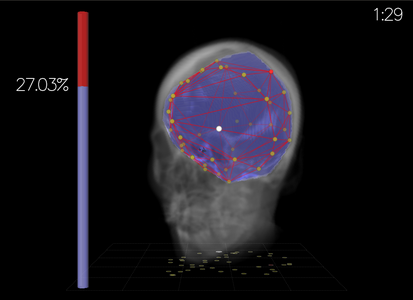





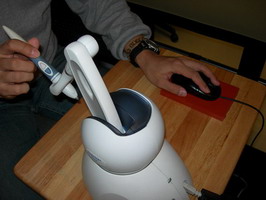

Task
Participants created a convex hull around a sub-volume target by
placing 3D mesh points. Error = excluded brain volume + included
non-brain volume.
Output Display
Compared perspective view (stereo) vs orthographic views.
Input Devices
Compared standard mouse with 3D devices: Depth Slider, Rockin’
Mouse, Wii remote, Phantom stylus, Space Navigator.
Results
Six participants (ages 23–38). Depth Slider offered advantages in
accuracy with bimanual control. Mouse remained accurate for precise
placement; orthographic slower but effective.